Martínez Baz






Iñigo Lasa holds a degree in Biology from the University of Navarra (Pamplona, 1988) and is the recipient of the Biological Sciences Extraordinary Award (Pamplona, 1988). He completed his doctoral thesis at the Severo Ochoa Molecular Biology Centre in the Autonomous University of Madrid (1992). Lasa worked as a post-doctoral researcher at the Max Planck Institute in Cologne (Germany, 1993) and the Institut Pasteur in Paris (1995-1997). In 2000, he was a visiting scientist at the Cold Spring Harbor Laboratory in New York.
In 1998, he was appointed Assistant Professor of Microbiology at the Public University of Navarra (UPNA); since 2008, after obtaining the national certification as a university professor, he has been Professor of Microbiology, specialising in Microbiology and Genetic Engineering. He has been the doctoral advisor of 13 students so far.
Iñigo Lasa has been leading researcher in 21 Spanish and European projects that produced more than 100 scientific papers published in international journals. At the domestic level, he was an adjunct at the National Agency for Project Assessment (ANEP) and is currently the manager of the Biotechnology area at the Sub-Directorate-General for Research Projects of the Ministry of Economy and Competitiveness.
In 2015, he was appointed Director of Navarrabiomed-Miguel Servet Foundation, while he continued leading the Microbial Pathogenesis Unit at Navarrabiomed.
Recently Iñigo Lasa has been appointed to membership in the European Academy of Microbiology, organization that includes leading European microbiologists in their fields and aims to promote excellence in microbiology.
The “Joint Action on integrating prevention, testing and link to care strategies across HIV, Viral Hepatitis, TB & STIs in Europe” (INTEGRATE) has the overall objective to increase Integrated early diagnosis and linkage to prevention and care of HIV, viral hepatitis, TB and STIs in EU Member States by 2020.
A number of tools have been developed to reduce transmission, optimize early diagnosis and linkage to care for one or more of these four diseases. INTEGRATE will map relevant existing tools for cross-linking. A peer-review process will identify which of these tools are complimentary or redundant for other disease(s), and which could be adapted or require further innovation.


The Microbial Pathogenesis Research Unit seeks to understand at the molecular level how pathogenic bacteria grow when adhering to the surface of medical devices and tissues, leading to infections that are resistant to antibiotics and therefore tend to become chronic. In order to understand this form of bacterial growth, known as biofilm, genetic engineering strategies are used, along with omics approaches, synthetic biology and animal models.
The ultimate goal is to identify the critical elements in biofilm formation in order to prevent biofilm from forming, eliminate already formed biofilm, improve existing treatments and favouring the formation of non-pathogenic bacteria biofilm for therapeutic purposes.
Lines of research:
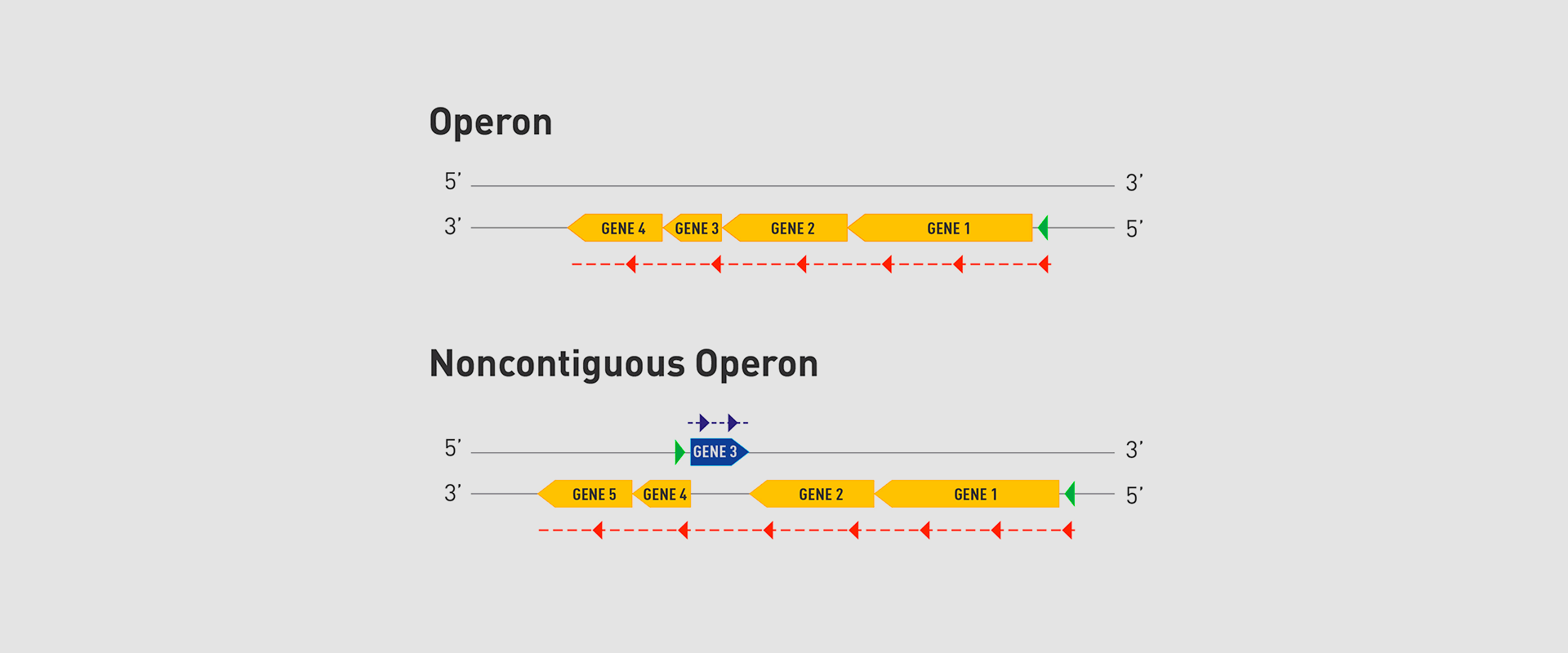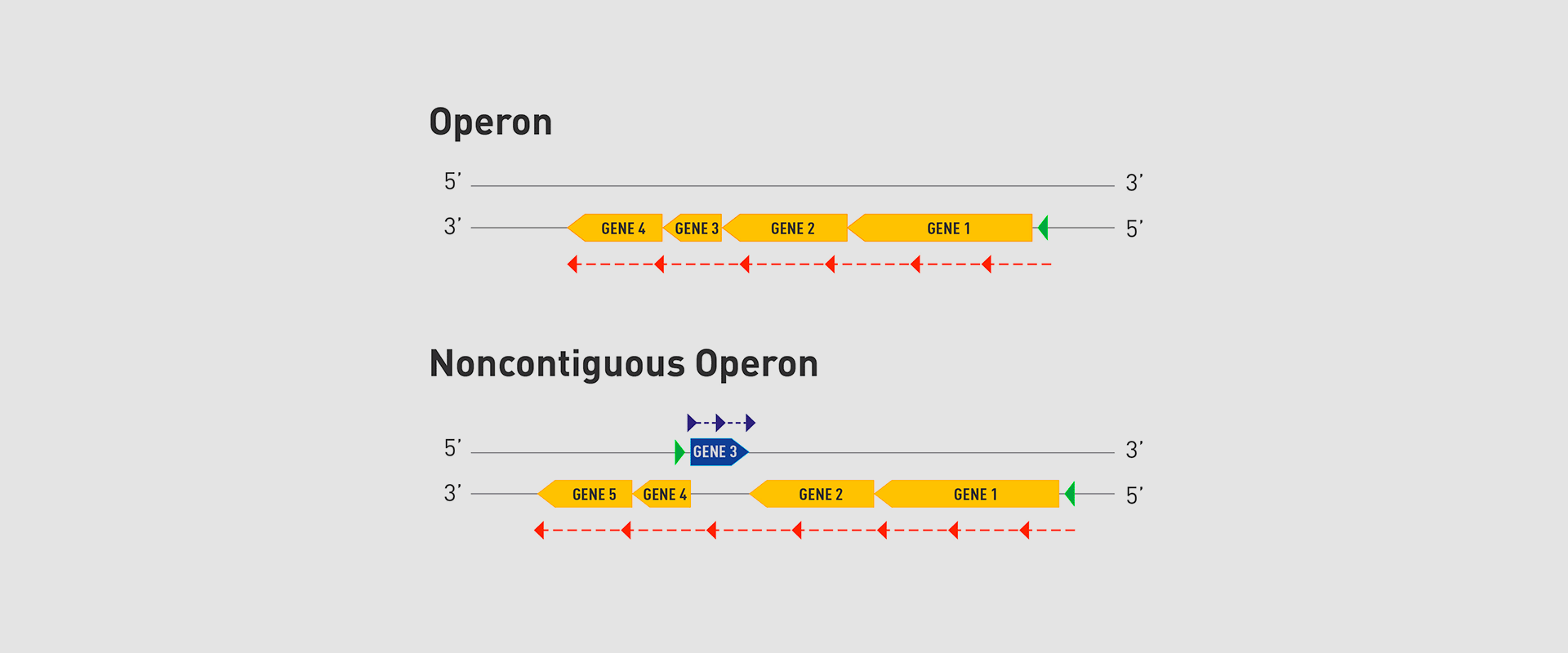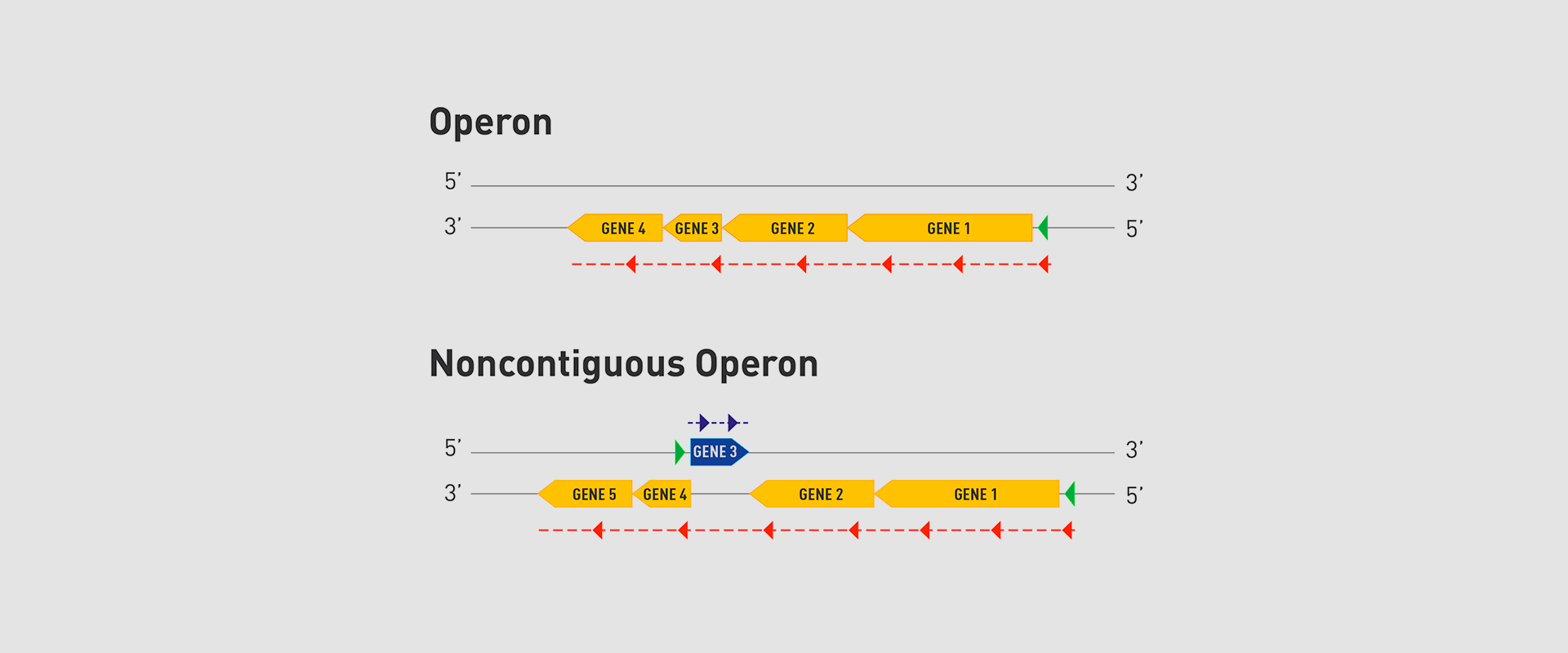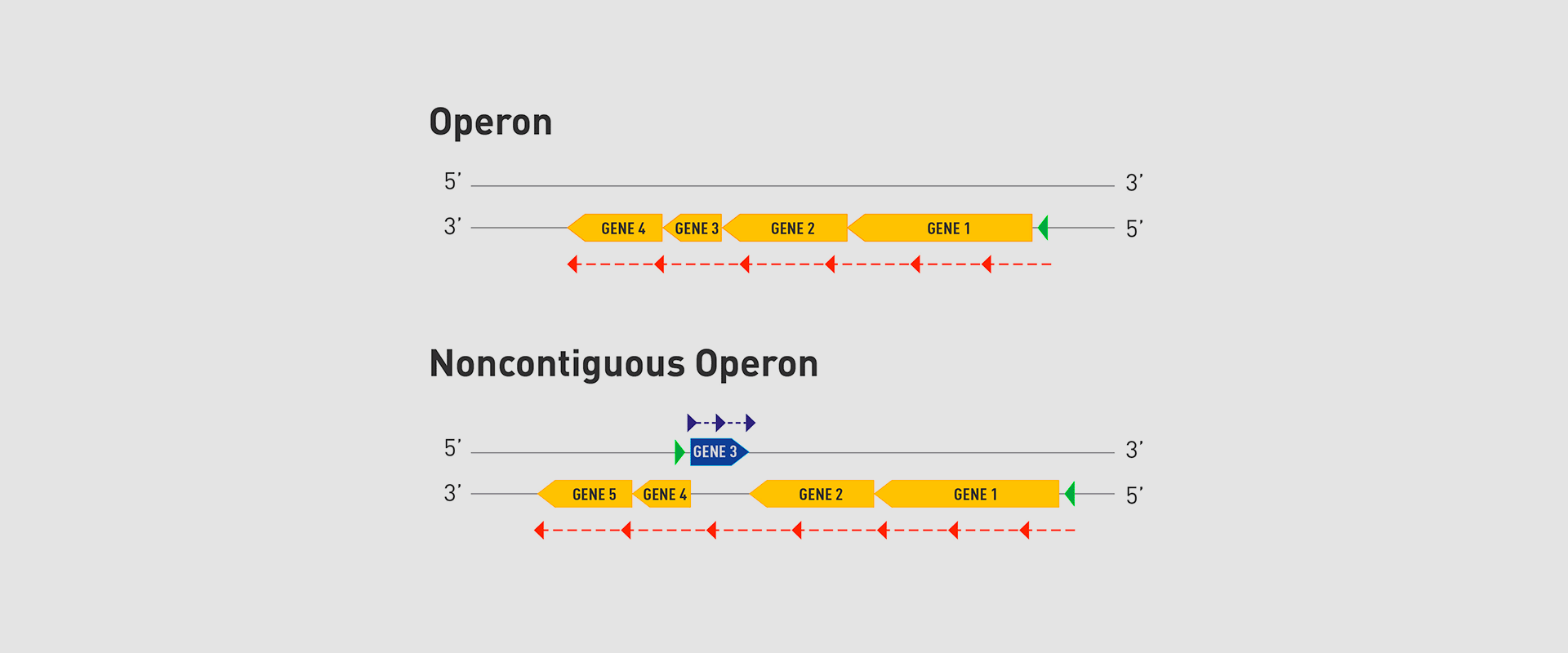
The Microbial Pathogenesis Unit of Navarrabiomed-Public University of Navarra (UPNA) has found a new genetic organisation in bacteria that helps better understand bacterial biology. The study of this genetic architecture was published in Proceedings of the National Academy of Sciences of the United States of America (PNAS).
Research background
In 1961, François Jacob and Jacques Monod discovered that bacteria group the genes that encode the proteins for a certain metabolic pathway in a single transcription unit (which they called ‘operon’). They won the Nobel Prize in Physiology or Medicine in 1965 for their discovery.
The bacteria they chose for their study was Escherichia coli, which normally lives in the intestines of healthy people; specifically, they studied the set of genes E. coli bacteria need to transport lactose (milk sugar) and break it down. E. coli only produces the three proteins it needs to digest lactose when the sugar is available. To simplify transcription regulation, the three genes involved are adjacent in the genome and under a single regulation system. Similar transcriptional regulation systems are found in other metabolic pathways in all bacteria.
Research at Navarrabiomed
In 2018, the team of researchers at Navarrabiomed coordinated by Iñigo Lasa Uzcudun, Head of the Microbial Pathogenesis Unit and Director of the biomedical research centre, described a new way genes are organised in bacteria. This regulation system has a higher level of regulation in operon structure, which the authors of the study named ‘non-contiguous operon’.
The bacterial model analysed has a group of four genes that are transcribed as a transcription unit despite the existence of a separate gene between the second and third genes that is transcribed in the opposite direction.
This transcriptional architecture results in an antisense transcript that acts as a mutual regulation system for the expression of the genes in the operon and the gene that produces this antisense transcript. Therefore, the concept of non-contiguous operon includes not only the genes transcribed from the same transcription unit but also overlapping genes whose expression is coordinated with that of the genes in the operon.
This finding deepens the understanding of bacterial biology and may trigger novel developments in the fields of synthetic biology and bacterial biotechnology.
The study was carried out as part of the scientific activity done at the Navarra Medical Research Institute (IdiSNA).

Un equipo científico del centro de investigación biomédica Navarrabiomed -centro mixto del Gobierno de Navarra y la Universidad Pública de Navarra (UPNA)- ha conseguido caracterizar el sistema sensorial que las bacterias utilizan entre otras cosas para multiplicarse en el cuerpo humano y causar infección.
El avance, que ha sido publicado por la revista científica Nature Communications y cuenta con financiación del Ministerio de Economía, Industria y Competitividad, permite comprender mejor cómo las bacterias se adaptan a las diferentes condiciones ambientales y posibilitará el desarrollo de antibióticos más específicos y eficaces.
El estudio ha contado con el liderazgo del doctor Iñigo Lasa, director de Navarrabiomed e investigador responsable del Grupo de Patogénesis Microbiana del centro. Asimismo, han colaborado investigadores del Instituto de Agrobiotecnología (UPNA-CSIC-Gobierno de Navarra), del Instituto de Biomedicina de Valencia (CSIC) y del Institute of Infection, Immunity and Inflammation, University of Glasgow.
Bacterias superresistentes
Actualmente, la aparición de bacterias farmacorresistentes, que no responden a tratamientos con antibióticos, constituye uno de los problemas sanitarios a escala mundial priorizados por la Organización Mundial de la Salud (OMS).
Las bacterias detectan, responden y se adaptan a los cambios en su entorno utilizando unos elementos sensoriales denominados sistemas de dos componentes. Este tipo de sistemas sensoriales están presentes en bacterias, hongos y plantas, pero no se encuentran en células animales. En el caso de las bacterias, regulan procesos celulares tan importantes como la virulencia o su propio crecimiento, lo que los convierte en dianas para el diseño de nuevas terapias antimicrobianas.
El objetivo del trabajo ha consistido en eliminar todos los sistemas de dos componentes, es decir el sistema sensorial completo, en Staphylococcus aureus, uno de los principales patógenos humanos según la OMS y, posteriormente, en la generación de una colección de bacterias cada una de las cuales contiene un único sistema de dos-componentes. Esta estrategia ha permitido simplificar una compleja red sensorial en cada uno de sus elementos para comprender cuál es la función individual de cada uno de los sistemas y la relación existente entre ellos.
Aplicación clínica de la investigación
En relación a la aplicación clínica, Iñigo Lasa apunta al desarrollo de nuevos antibióticos más específicos. “El hecho de que los sistemas de dos componentes estén presentes en todas las bacterias patógenas y no en las células de nuestro organismo nos puede permitir desarrollar fármacos que bloqueen estos sistemas, evitando así el desarrollo de la bacteria durante la infección, sin causar ningún efecto secundario sobre nuestras células”.
En este sentido, las bacterias generadas en este estudio han sido patentadas y actualmente el equipo analiza diversos compuestos marinos que puedan incorporarse en el tratamiento y control de infecciones en la práctica clínica.
La investigación forma parte de la actividad científica del Instituto de Investigación Sanitaria de Navarra (IdiSNA), agrupación público-privada para el fomento de la investigación biomédica en la Comunidad Foral y de la que son miembros Navarrabiomed y la UPNA.

Navarrabiomed - Centro de investigación biomédica
Complejo Hospitalario de Navarra, edificio de investigación.
Calle Irunlarrea, 3. 31008 Pamplona, Navarra, España.